Abstract
BACKGROUND: The prognostic and therapeutic impact of TP53-mutated (TP53mut) myeloid neoplasm (MN) is an area of intense interest, with the 5th edition of the World Health Organization, the International Consensus Classification, and the European LeukemiaNet guidelines, all recommending major changes MN classification based on TP53mut. However, studies driving these changes predominantly included de novo MDS (Bernard et al. Nat Med 2020), AML and high risk MDS treated with intensive chemotherapy (Grob et al. Blood 2022), or MN with complex karyotype (CK, Weinberg et al. Blood Adv 2022). Therefore, the significance of these findings as they relate to t-MN is not known.
METHODS: We analyzed therapy-related myeloid neoplasm (t-MN) (n=377) patients evaluated at Mayo Clinic, Rochester and South Australian MDS Registry with clinical, integrated genomic profiling, and long-term follow-up to characterize the genomic profile and outcome of TP53mut t-MN.
RESULTS: Our cohort of 377 t-MN included t-MDS (n=245, 65%) and t-AML (n=132, 35%), with a median age at t-MN diagnosis being 65.7 years. 184 TP53mut at variant allele frequency (VAF) ≥2% were detected in 131 (34.74%) patients. Median overall survival (OS) of TP53mut VAF <10% (n=14) was similar to wild-type TP53 (TP53wt), hence further analysis was restricted to TP53mut with VAF >10% (n=111). TP53mut patients without VAF (n=6) were not included for further analysis.
Genomic instability was highly evident in TP53mut t-MN compared to TP53wt t-MN. CK, monosomal karyotype (MK), chromosome 5q deletion, and marker chromosomes were enriched in TP53mut t-MN. In contrast, driver oncogenic gene mutations such as IDH2, SRSF2, RAS, and TET2 were less frequent in TP53mut. TP53mut also dictated the clinical presentation, longer latency (interval between primary cancer and t-MN diagnosis), and inferior OS compared to TP53wt (8.2 vs. 19.4 months; P <0.001). On multivariate Cox regression analyses, TP53mut was associated with inferior OS (HR 1.43 95% CI 1.01-2; P =0.03) independent of CK and bone marrow blast %.
A significantly higher proportion of CK patients had TP53mut compared to non-CK patients (74.4% vs. 5.4%, P <0.001). Even within the CK cohort, TP53mut was associated with structural genomic instability, with a higher proportion of TP53mut harbouring MK (P =0.013), marker chromosome (P =0.033), deletion 5q (P <0.001), abnormal chromosome 17 (P =0.001), and other structural chromosomal aberrancies compared to TP53wt-CK. In contrast to the predominance of structural chromosomal aberrancies in the TP53mut-CK cohort, mutations driving MN were sparse in TP53mut-CK compared to TP53wt-CK. Finally, TP53mut-CK was associated with significantly poor survival compared to the TP53wt-CK (8.2 vs. 17 months; P <0.001).
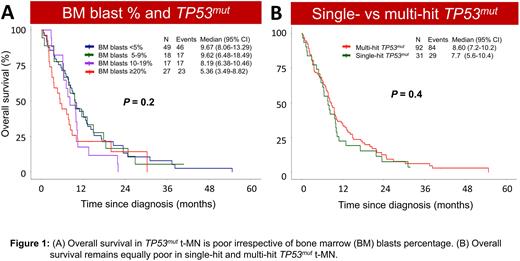
Though bone marrow (BM) % predicted poor outcome in the whole t-MN cohort, it failed to predict outcome in TP53mut t-MN patients. We compared the clinical and molecular profiling of TP53mut t-MDS (<5%, 5-9%, 10-19% blasts) and t-AML (≥20% blasts) at t-MN diagnosis. Genomic instability, TP53mut allelic status, number of TP53mut, and the clone size were similar across all four cohorts. Remarkably, the survival of TP53mut patients were equally dismal irrespective of blast percentage (P=0.2, Figure 1A). Together, these findings suggest that TP53mut t-MN represent a homogenous group irrespective of BM blast percentage.
Integrating the mutation, conventional cytogenetic, copy number variation, FISH, and SNP array analysis, TP53mut were classified as single- or multi-hit (Grob et al. Blood 2022). In total, 31 (25.2%) of the 123 patients with TP53mut (n=111) and/or loss of TP53 (n=12) locus were considered single-hit, and 92 (74.8%) were considered as multi-hit. In contrast to prior reports, we did not observe enrichment of genomic instability, or differences in degree of cytopenia, bone marrow blast % and t-MN phenotype in multi- vs single-hit TP53mut patients. Importantly, no OS differences were observed when TP53mut were stratified by type of TP53mut (truncation vs. missense), site of missense mutation (hot- vs. non-hot spot) and single- vs multi-hit (Figure 1B).
CONCLUSION: The classification of t-MN based on TP53mut status is clinically and biological relevant, irrespective of blast % or single- or multiple-hit status. TP53mut t-MN is a molecularly defined unique group enriched for genetic instability and is associated with extremely poor outcomes.
Disclosures
Shah:Astellas: Research Funding; Celgene: Research Funding; Marker Therapeutics: Research Funding. Sharplin:Novartis: Other: Travel and conference funding; Kite: Honoraria. Al-Kali:Astex: Other: research support to institution. Ross:Takeda: Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Keros: Consultancy; Celgene: Research Funding; BMS: Honoraria. Yeung:Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees; Amgen: Honoraria; Pfizer: Honoraria; BMS: Honoraria, Research Funding. Shanmuganathan:Novartis: Honoraria; Amgen: Other: Meeting sponsorship. Litzow:Abbvie: Research Funding; Amgen: Research Funding; Astellas: Research Funding; Novartis: Research Funding; Syndax: Research Funding; Jazz: Consultancy; Actinium: Research Funding; Pluristem: Research Funding; Biosight: Other: Data Monitoring Board. Mangaonkar:Bristol Myers Squibb: Research Funding. Patnaik:Kura Oncology, Stemline Therapeutics: Research Funding. Hiwase:Novartis: Speakers Bureau; BMS: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; AbbVie: Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal